| D-Strap DNA Sequencing Chemistry
We have developed D-Strap chemistry primarily for direct genomic DNA sequencing. D-Strap allows initiating and continuing genomic DNA sequencing projects without shotgun libraries. It increases the robustness of primer walk technology on genomic DNA templates and is especially beneficial when several related DNAs are sequenced simultaneously. D-Strap uses evolutionary conserved sequence elements in design of Fimers for sequencing genomic DNA templates.
Most of D-Strap Fimers contain degenerate bases, whereby they also employ the advanced Fimer chemistry to adjust melting temperatures for optimal primer annealing and prevent formation of DNA secondary structures in sequencing reactions.
The use of D-Strap Fimers is especially beneficial for direct sequencing genomic DNAs from related organisms, as D-Strap Fimers can accommodate multiple degenerate bases.
D-Strap Sequencing BACs with Primate Genes
|
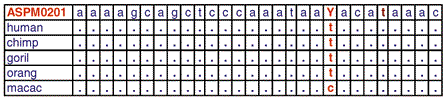
Fimer ASPM0201 is designed for human ASPM gene (top table). It contains one degenerate base (Y). The same Fimer ASPM0201 was used for direct sequencing the ASPM genes from chimpanzee, gorilla, orangutan, and macaque captured in BACs.
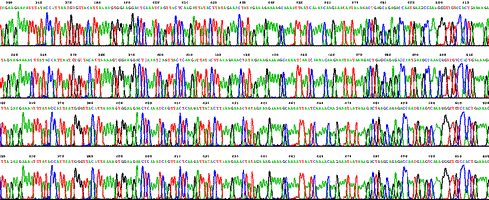
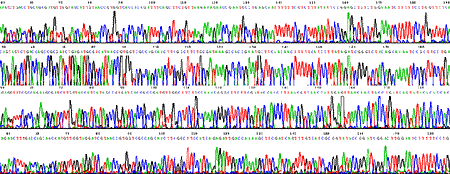
Using a set of D-Strap Fimers (0 to 3 degenerate bases) we were able to sequence complete genes from six primates. The example shows parts of 1000 bp long high quality traces. Sequences of ASPM genes from chimp, gorilla, orangutan and macaque obtained with D-Strap technology do not contain sequencing errors, cloning or PCR artifacts.
|
D-Strap Sequencing Bacterial Genomes
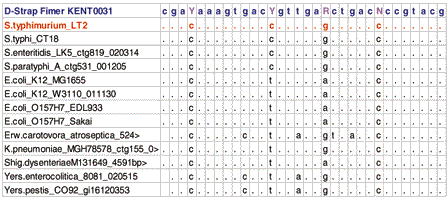 |
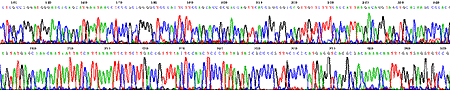
Fimer KENT0031 was designed from the alignment of Enteric bacteria. It had four degenerate bases (Y,Y,R,N). Fimer KENT0031 was used for direct sequencing both from Salmonella typhimurium strain LT2 (top trace), E. coli B (bottom trace) genomic DNAs.
|

Fimer DLEP0022 is designed from the alignment of Lactic Acid bacteria with three degenerate bases (Y,S,Y). The Fimer was used for direct sequencing from 15 LAB genomic DNA samples. The trace for Lactococcus lactis subsp. cremoris strain SK11 is shown. |
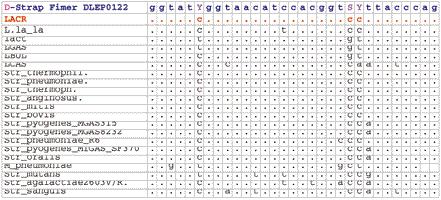
|
D-Strap Sequencing of Bacteriophage Genomic DNA
The diversity of DNA sequences of bacterophages is much higher than that of bacterial or eukariotic species. To test the limits of D-Strap chemistry we have designed and validated a
set of D-Strap Fimers to work on T4-like bacteriophage
genomic DNAs that target weakly conserved genes. |
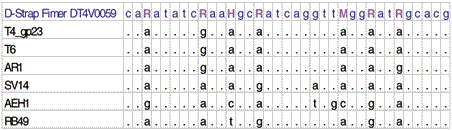
Fimer DT4V0059 was designed using alignment of T4-like bacteriophages with seven degenerate bases (R,R,H,R,M,R,R). The traces obtained by direct sequencing genomic DNAs from bacteriophages T4, RB49, AEH1, and 31 with Fimer DT4V0059 are shown.
|
Covered by U.S. Patents 5,427,928, 5,656,463, 5,902,879, 6,548,251. Patents pending. Fidelase, ThermoFidelase, D-Strap and Fimer are trademarks of Fidelity Systems. |






