Direct Sequencing of Microbial Genomic DNA
|
Direct genomic DNA sequencing eliminates the need in subcloning and production of many shotgun libraries, minimizes the number of sequencing reactions, and dramatically accelerates the assembly of complete sequences.
A core component of the procedure is the use of genomic DNA as a template in a robust sequencing reaction. The addition of ThermoFidelase with a unique combination of topoisomerase and DNA binding activities shortens the cycles of denaturation and primer annealing. The dramatic increase in specificity, quality, and yield of priming on large templates is achieved by using Fimers instead of regular primers. Fimers allow for multiplying the number of thermal cycles in sequencing reactions, which greatly increases the amplification of labeled products and the sequencing signals.
The advantages of direct genomic sequencing include elimination of cloning artifacts and cross-contamination of libraries or PCR reactions. This is extremely important for production sequencing of closely related organisms, as it provides non-biased complete coverage of the genomes with low number of redundant sequencing reactions and results in significant savings on data processing.
Archaebacterial Genomic DNA Bacterial Genomic DNA 1 Bacterial Genomic DNA 2 Direct sequencing off microbial DNA
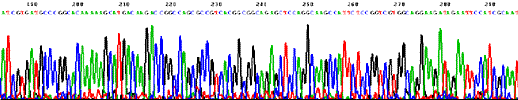
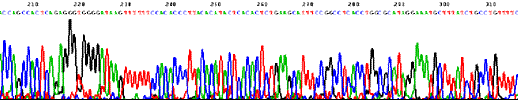
Direct sequencing of E. coli genome (4.6 Mb) 1.4 Mb circular chromosome of M. kandleri strain TAG11 was sequenced using D-Strap technology.

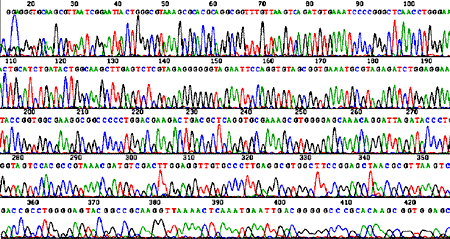
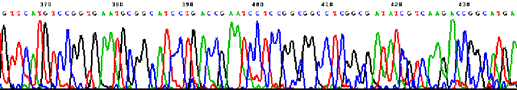
 Sequencing trace from Novosphingobium aromaticivorans DSM 12444 (one 4 Mb circular chromosome).  Sequencing trace from Klebsiella pneumoniae MCG 78578 (one 6 Mb circular chromosome). 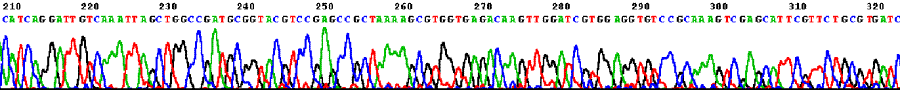 Sequencing trace from Pseudomonas syringae pathovar maculicola 6.4 Mb genomic DNA.
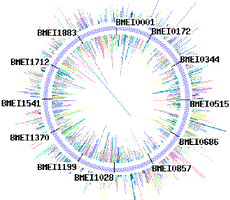
Sequencing traces from each chromosome of Brucella melitensis strain 16M (two circular chromosomes: 2,117,144 and 1,177,787 bp).
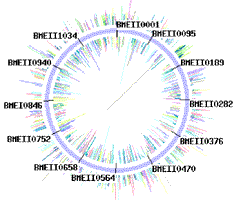
Sequencing traces from each chromosome of Agrobacterium tumefaciens strain C58 (one 2,841,581 bp circular chromosome and one 2,074,782 bp linear chromosome).
|